Case Studies
Case Study: University of Texas Medical Branch, Biomolecular Resource Facility
The University of Texas Medical Branch, Biomolecular Resource Facility is one of only seven sites within the USA that receive multi-million dollar state funding by the US Department of Health & Human Services.

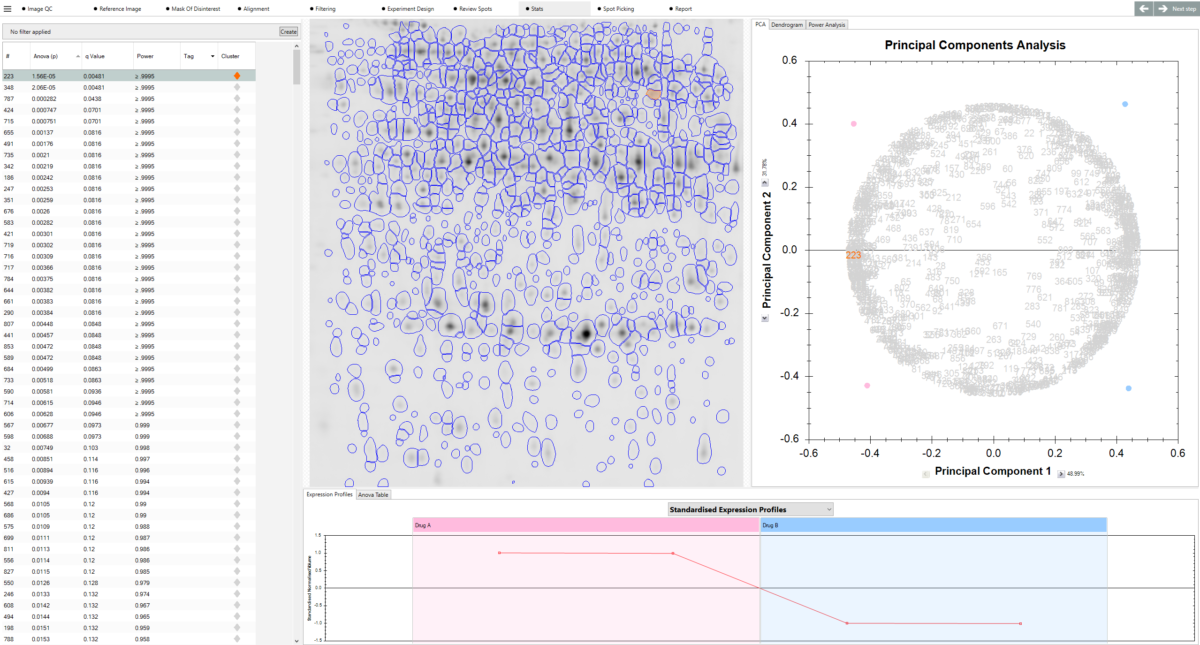
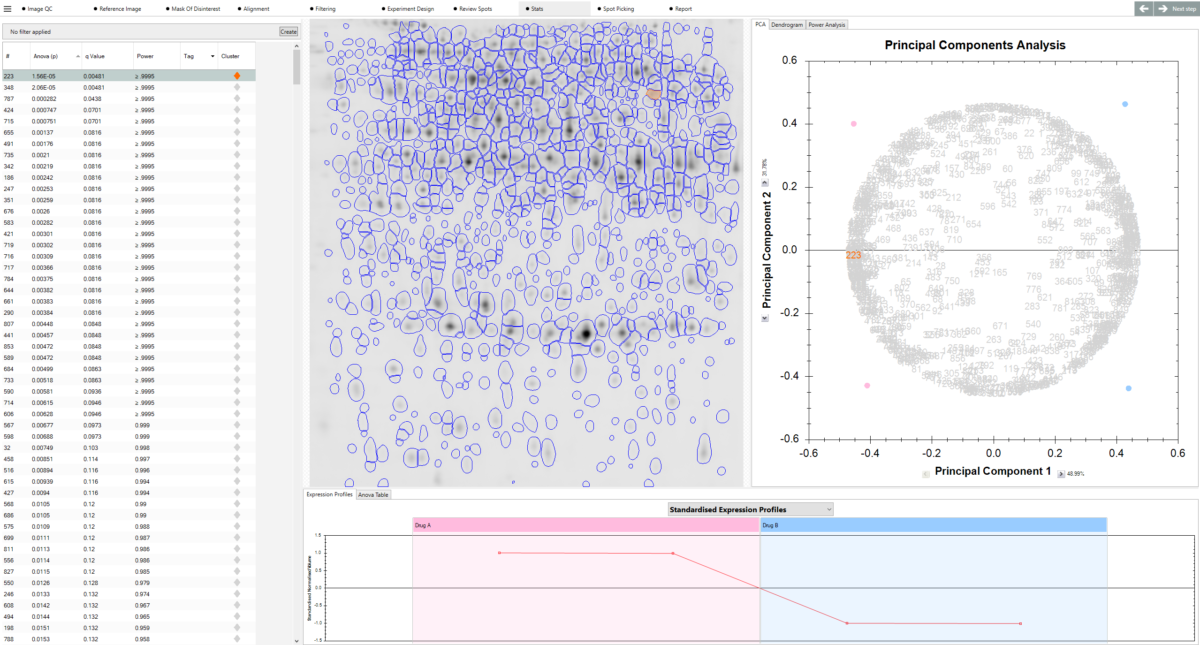
“The fundamental concept (image alignment) of SameSpots permits us greatly increased confidence in quantification, and this is enhanced when performed on gels obtained by our saturation fluorescence strategy. The statistical package further enhances confidence in our quantitative studies. In addition SameSpots has increased our throughput – with the demand we experience, this results in tremendous savings in manpower and expense over a year. With the proper tools and attention to detail proteomics can reap enormous benefits whatever your field of biological study. SameSpots is one of the tools that are indisputably required — providing the image and subsequent statistical analyses that are critical to a conclusive and compelling proteomics study.”
John E. Wiktorowicz, Ph.D. Associate Professor, Dept. Biochemistry & Molecular Biology Director, Proteomics Section
Case Study: Istituto di Ricerche Farmacologiche “Mario Negri”
Founded in 1963, The Istituto di Ricerche Farmacologiche “Mario Negri” is a non-profit independent scientific organisation for biomedical research and education. “In the Environmental Health Sciences Department we investigate the effect of environmental factors on human health, focussing on the survey of environmental contaminants, the assessment of human exposure with related health.

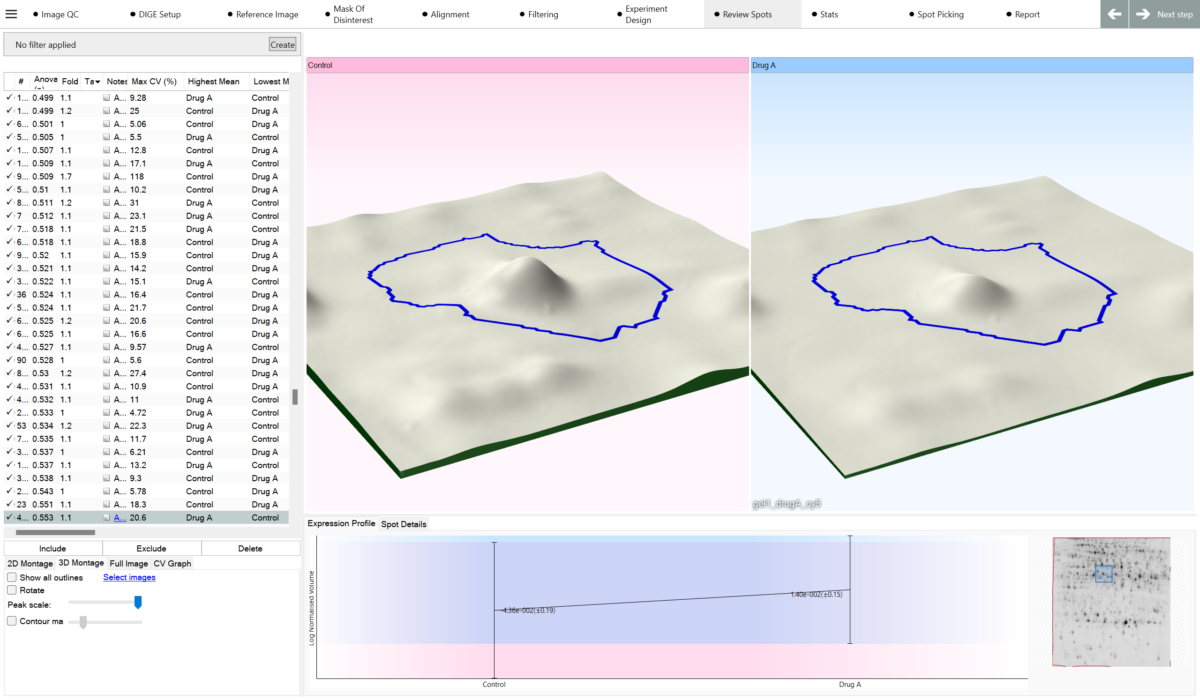
“We have been using SameSpots in our research since January 2007. The software has drastically simplified our proteomics workflow and opens up new avenues for the exploration of the data obtained”
“Since using SameSpots the speed of our analysis has been drastically improved. We align our gels accurately within 5 mins per gel and then run the SameSpots analysis. We don’t usually perform any correction of spot detection which results in fast, robust and very reproducible analysis. This has an immediate positive effect on the quality and confidence in our results.’’
Dr Roberta Pastorelli, Head, Protein and Gene Biomarkers Unit Laboratory of Molecular Toxicology
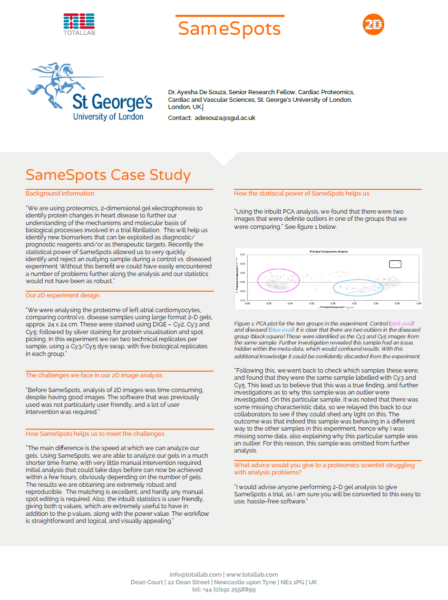
Case Study: Cardiovascular Sciences Research Centre, St. George’s University of London
The Cardiovascular Sciences Research Centre, St. George’s University of London, is a multidisciplinary group which amalgamates clinical, surgical and basic science research expertise. The mission of the Research Centre is to understand mechanisms of cardiovascular disease.

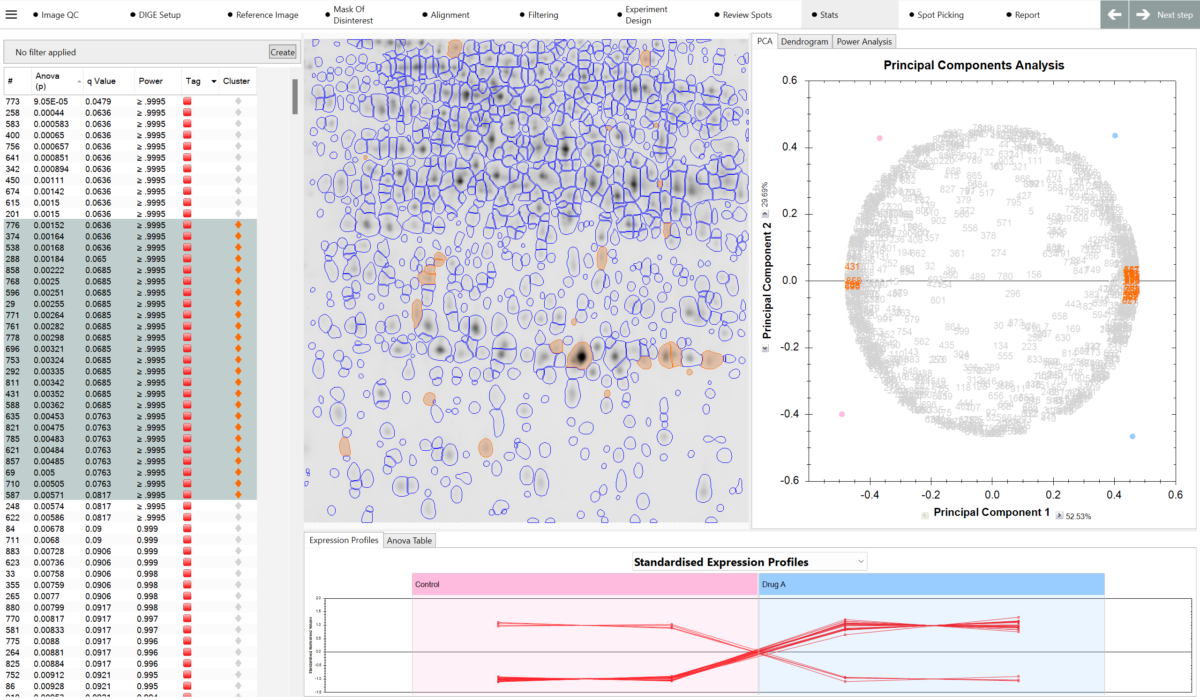
“The main difference is the speed at which we can analyze our gels. Using SameSpots, we are able to analyze our gels in a much shorter time frame, with very little manual intervention required. Initial analysis that could take days before can now be achieved within a few hours, obviously depending on the number of gels. The results we are obtaining are extremely robust and reproducible. The matching is excellent, and hardly any manual spot editing is required. Also, the inbuilt statistics is user friendly, giving both q values, which are extremely useful to have in addition to the p values, along with the power value. The workflow is straightforward and logical, and visually appealing.”
Dr. Ayesha De Souza, Senior Research Fellow, Cardiac Proteomics, Cardiac and Vascular Sciences, St. George’s University of London, London, UK.
Case Study: Centre Paul Papin
The Centre Paul Papin is a hospital specialized in cancer treatment and cancer research. The Centre Paul Papin collaborates with the Plateforme technologique d’Angers which offers, among other things, services in proteomics to public laboratories and industry.

“With our previous software, we experienced three major problems. Firstly, despite the time we spent disposing of many landmarks on the master gel, the software made many mistakes in alignment. Secondly, when a spot was missing on the gel master, it was not detected and analysed. Finally, the software was not friendly to use and the analysis was laborious.”
“SameSpots is so convivial and intuitive that it is a real pleasure to work with. The image alignment step makes us feel secure that we have one hundred percent matching and the additional statistical tools are perfectly designed for biologists who are not specialized in statistics. The development and improvement of SameSpots has been pleasing.”
Prof Catherine Guette, Proteomics platform leader, Laboratoire d’oncopharmacologie, Nice, France